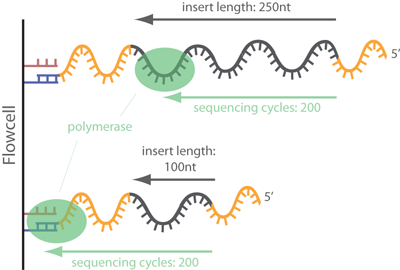
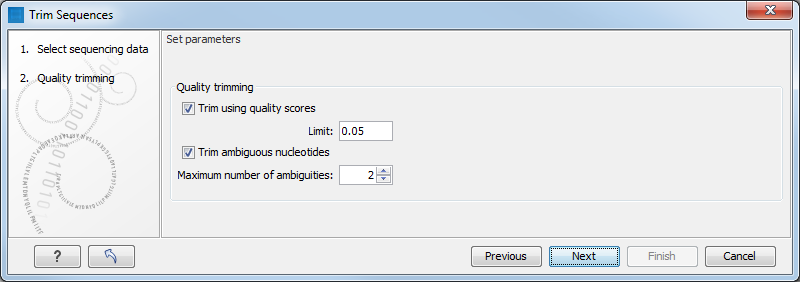
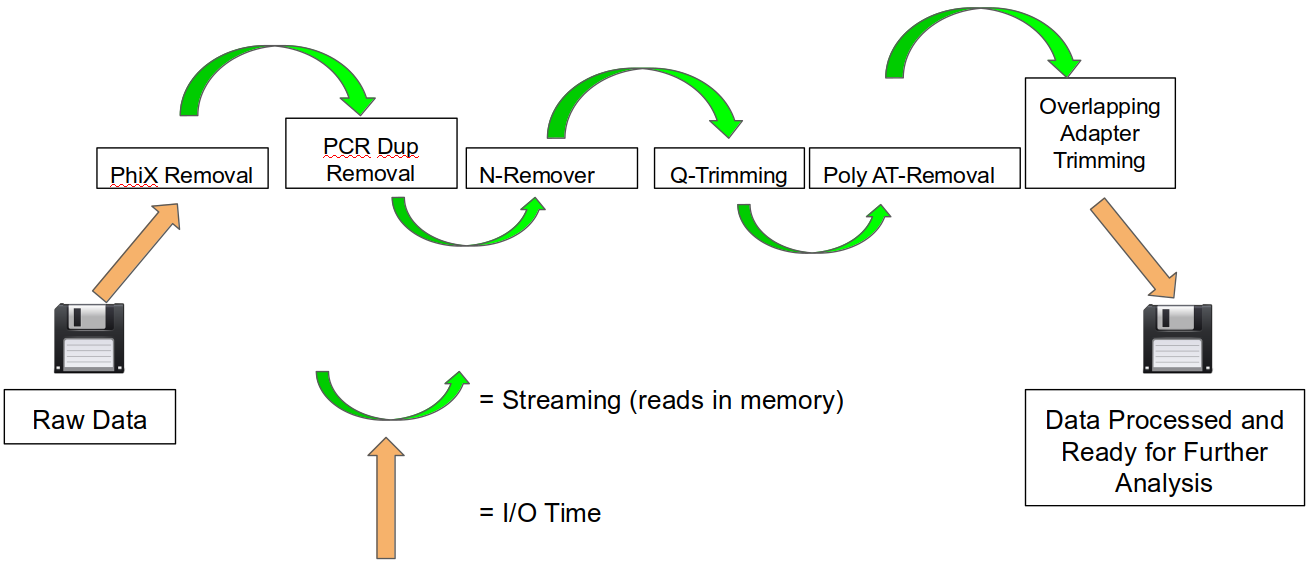
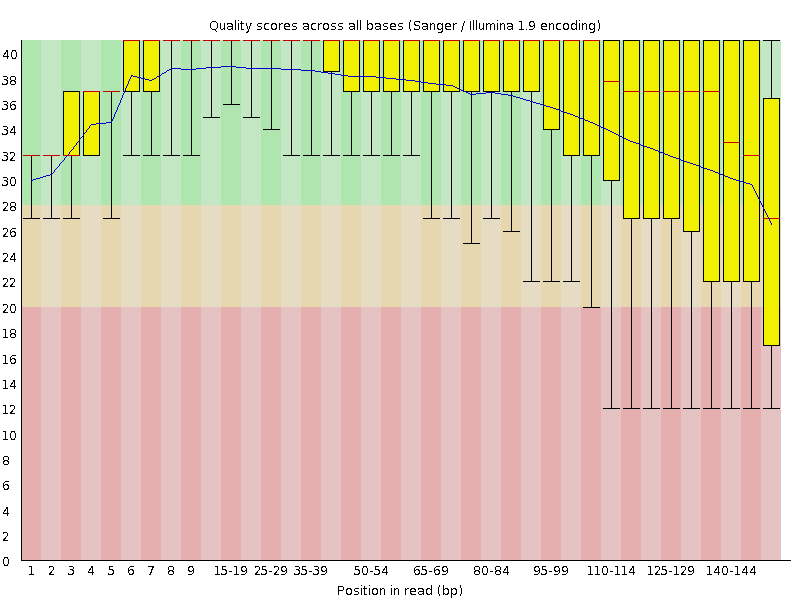
SeqPurge: highly-sensitive adapter trimming for paired-end NGS data – topic of research paper in Clinical medicine. Download scholarly article PDF and read for free on CyberLeninka open science hub.
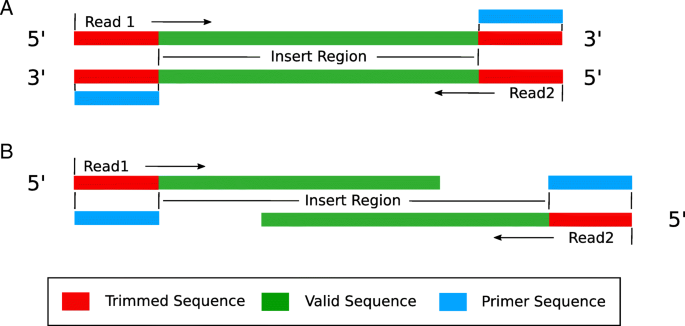
pTrimmer: An efficient tool to trim primers of multiplex deep sequencing data | BMC Bioinformatics | Full Text

Help with trimming SE reads using dada2- what would you suggest as the best trim/trunc parameters? : r/bioinformatics

A Bioinformatics Whole-Genome Sequencing Workflow for Clinical Mycobacterium tuberculosis Complex Isolate Analysis, Validated Using a Reference Collection Extensively Characterized with Conventional Methods and In Silico Approaches | Journal of ...

SeqPurge: highly-sensitive adapter trimming for paired-end NGS data | BMC Bioinformatics | Full Text
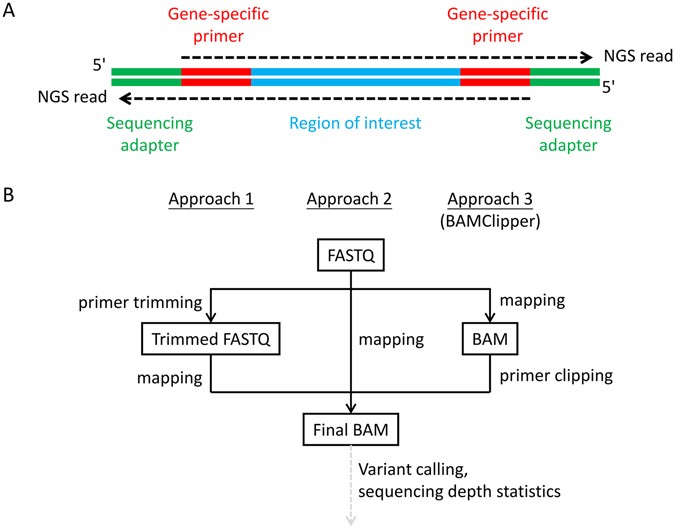
BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing | Scientific Reports

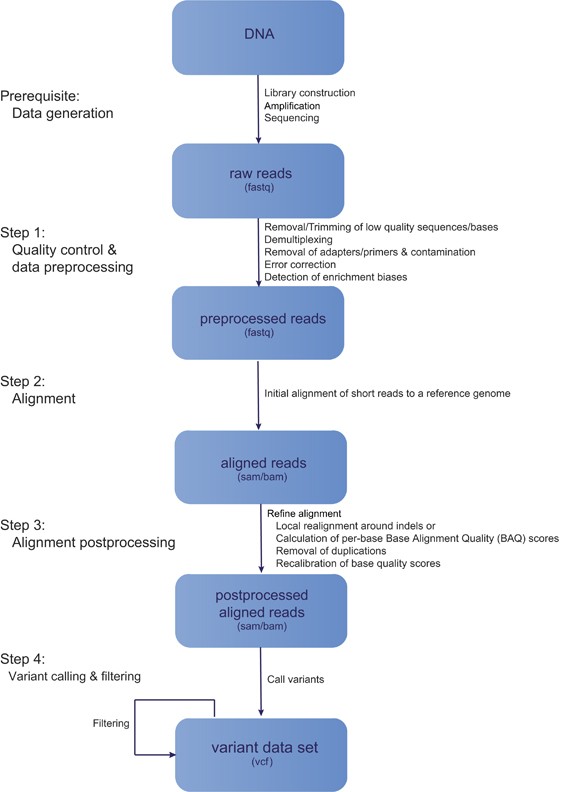
Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines: A Joint Recommendation of the Association for Molecular Pathology and the College of American Pathologists - ScienceDirect
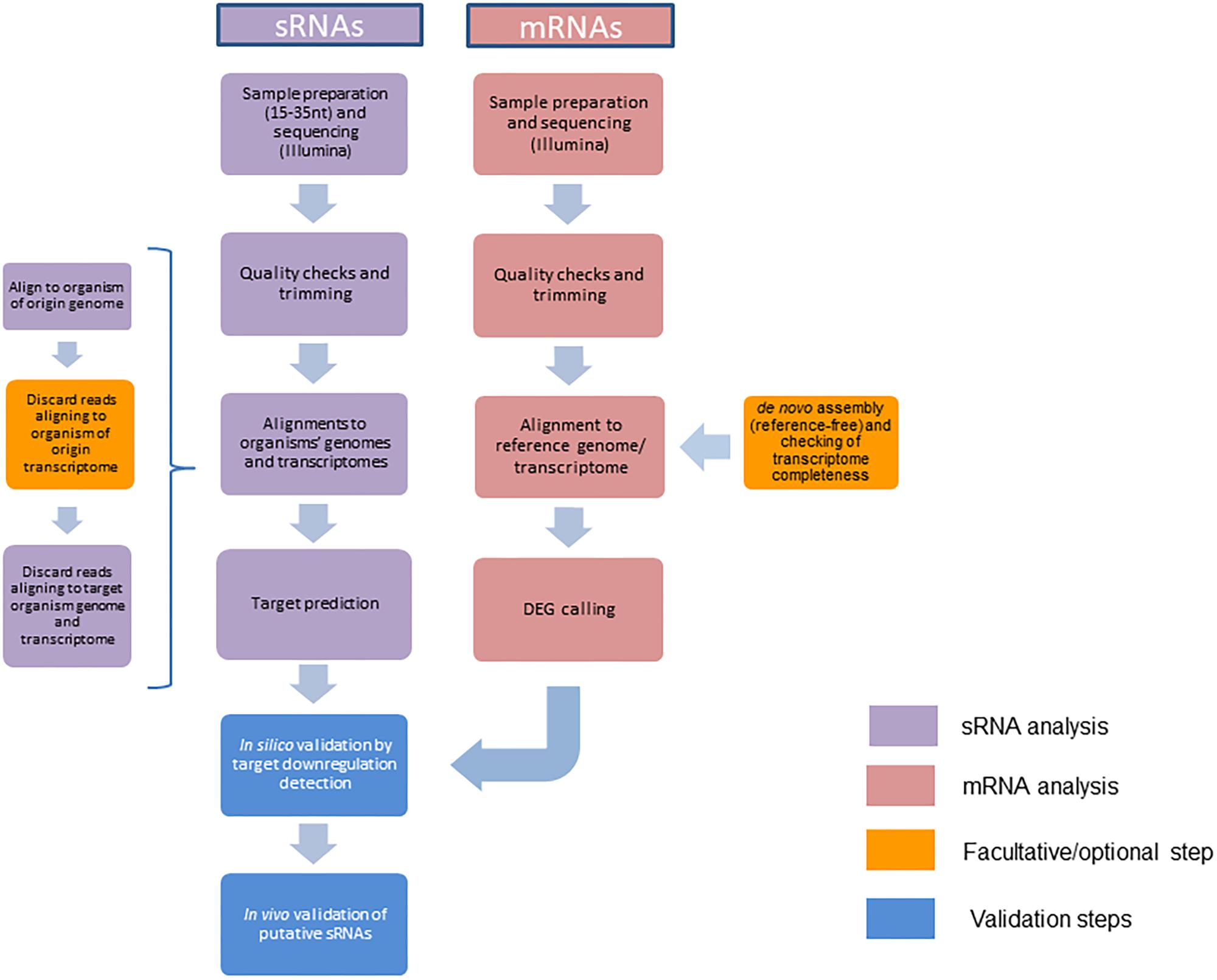
Frontiers | A Bioinformatics Pipeline for the Analysis and Target Prediction of RNA Effectors in Bidirectional Communication During Plant–Microbe Interactions
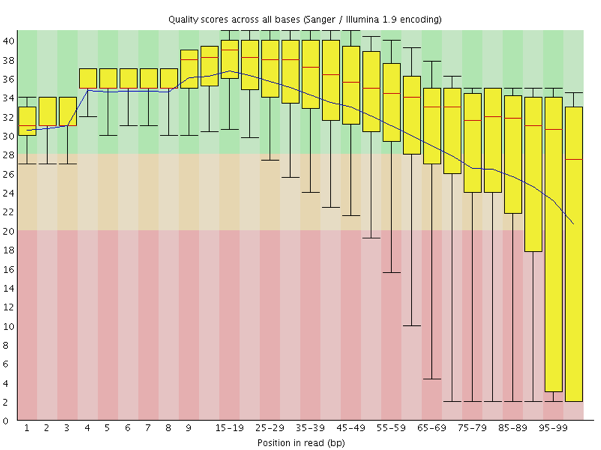
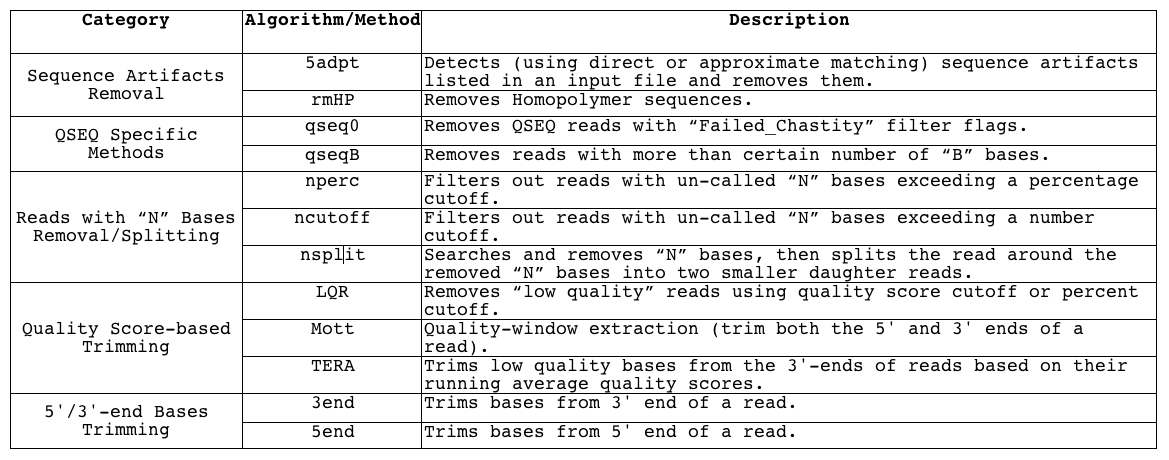
FASTQ Quality Assurance Tools - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis
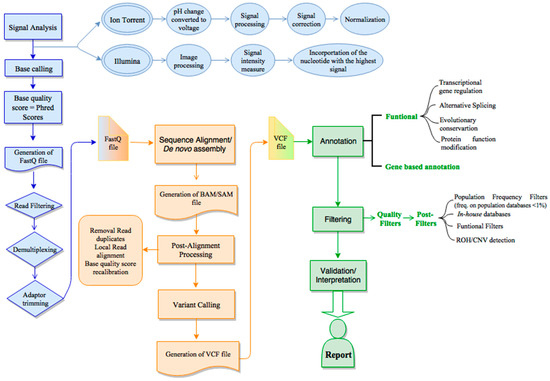
JCM | Free Full-Text | Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics
![SECAPR—a bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments [PeerJ] SECAPR—a bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2018/5175/1/fig-1-full.png)