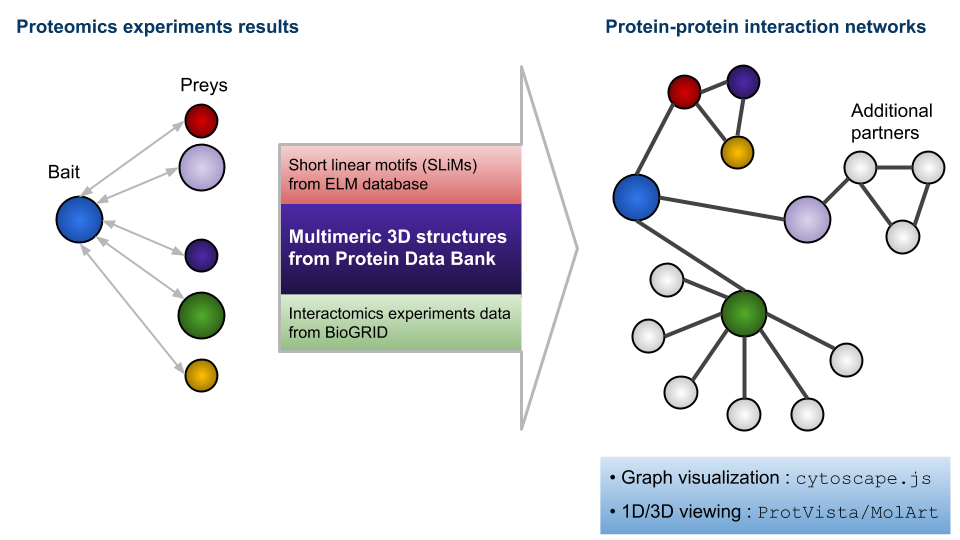
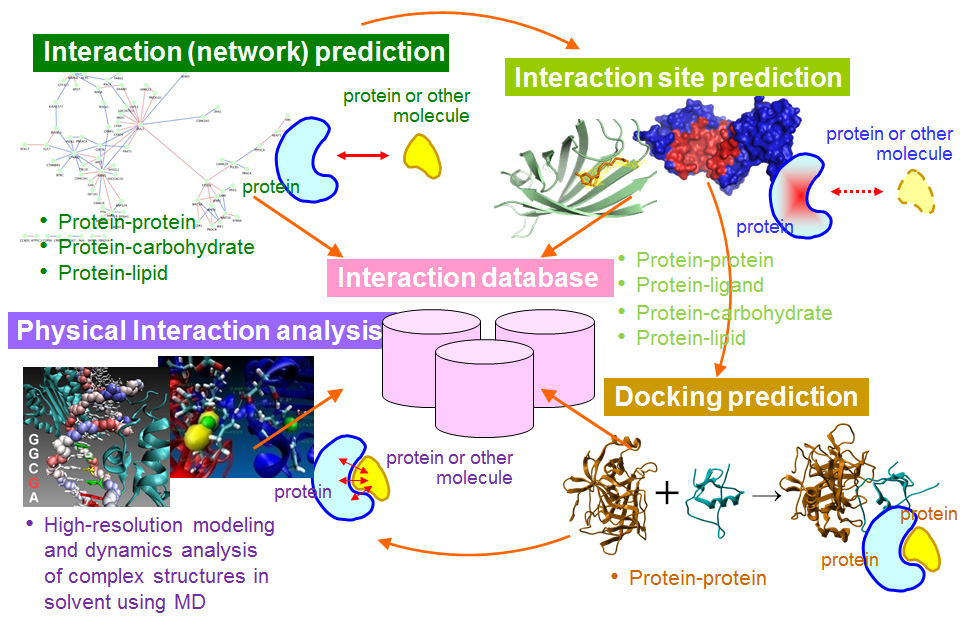
In silico-prediction of protein–protein interactions network about MAPKs and PP2Cs reveals a novel docking site variants in Brachypodium distachyon | Scientific Reports

Docking-based identification of small-molecule binding sites at protein- protein interfaces - Computational and Structural Biotechnology Journal

Protein ^ protein interaction prediction. PPIs can be predicted based... | Download Scientific Diagram

Predicting Protein-Protein Interactions from Matrix-Based Protein Sequence Using Convolution Neural Network and Feature-Selective Rotation Forest | Scientific Reports
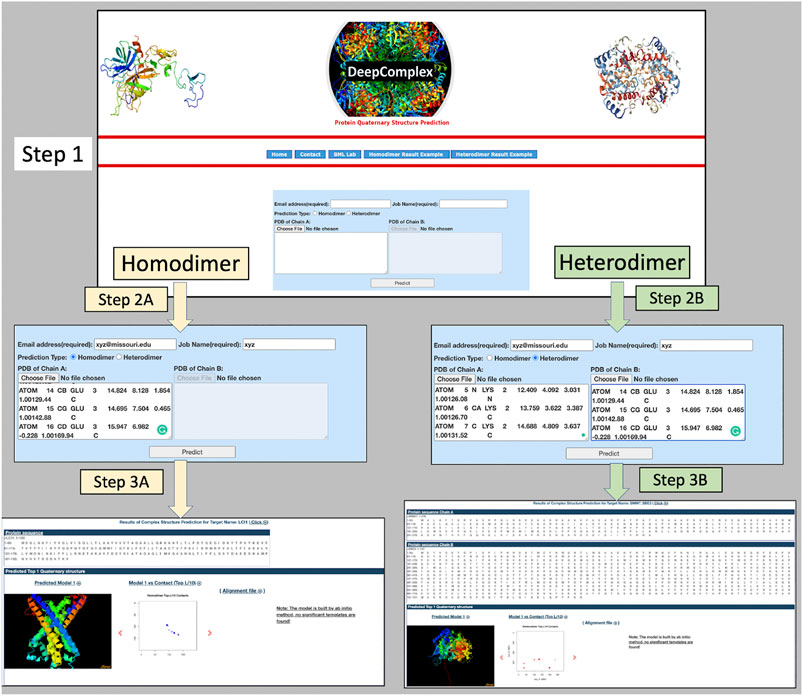
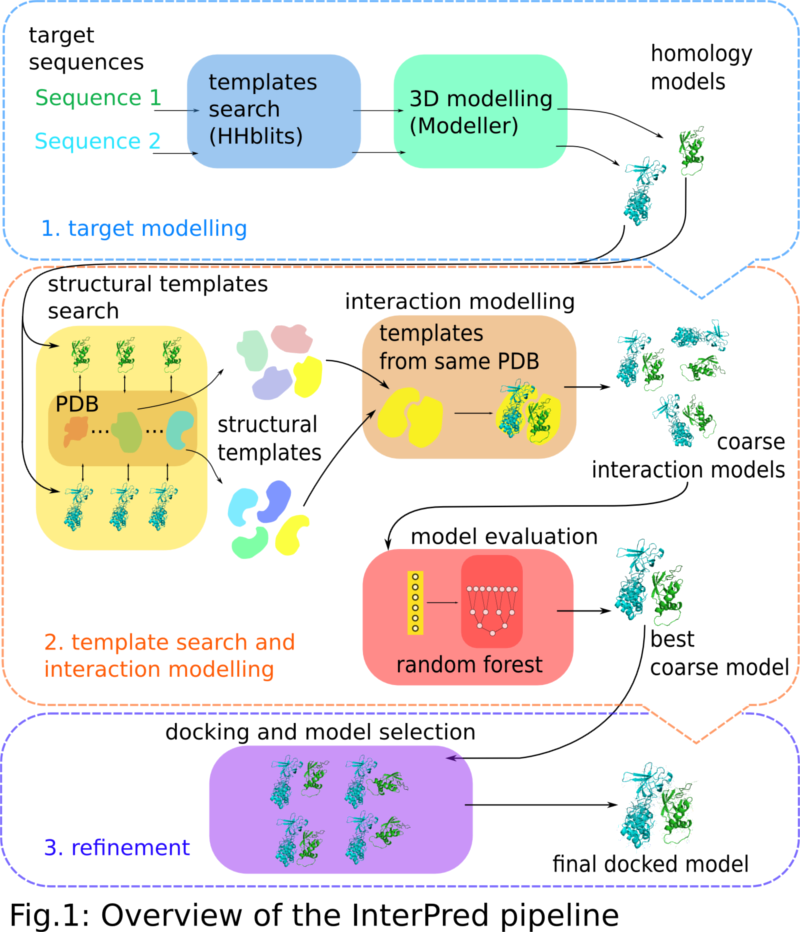
Frontiers | DeepComplex: A Web Server of Predicting Protein Complex Structures by Deep Learning Inter-chain Contact Prediction and Distance-Based Modelling
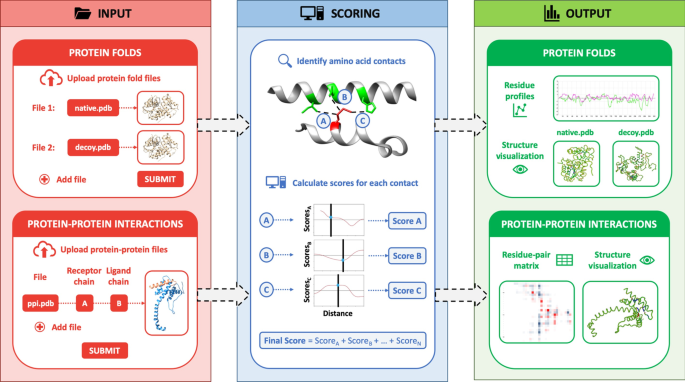
SPServer: split-statistical potentials for the analysis of protein structures and protein–protein interactions | BMC Bioinformatics | Full Text

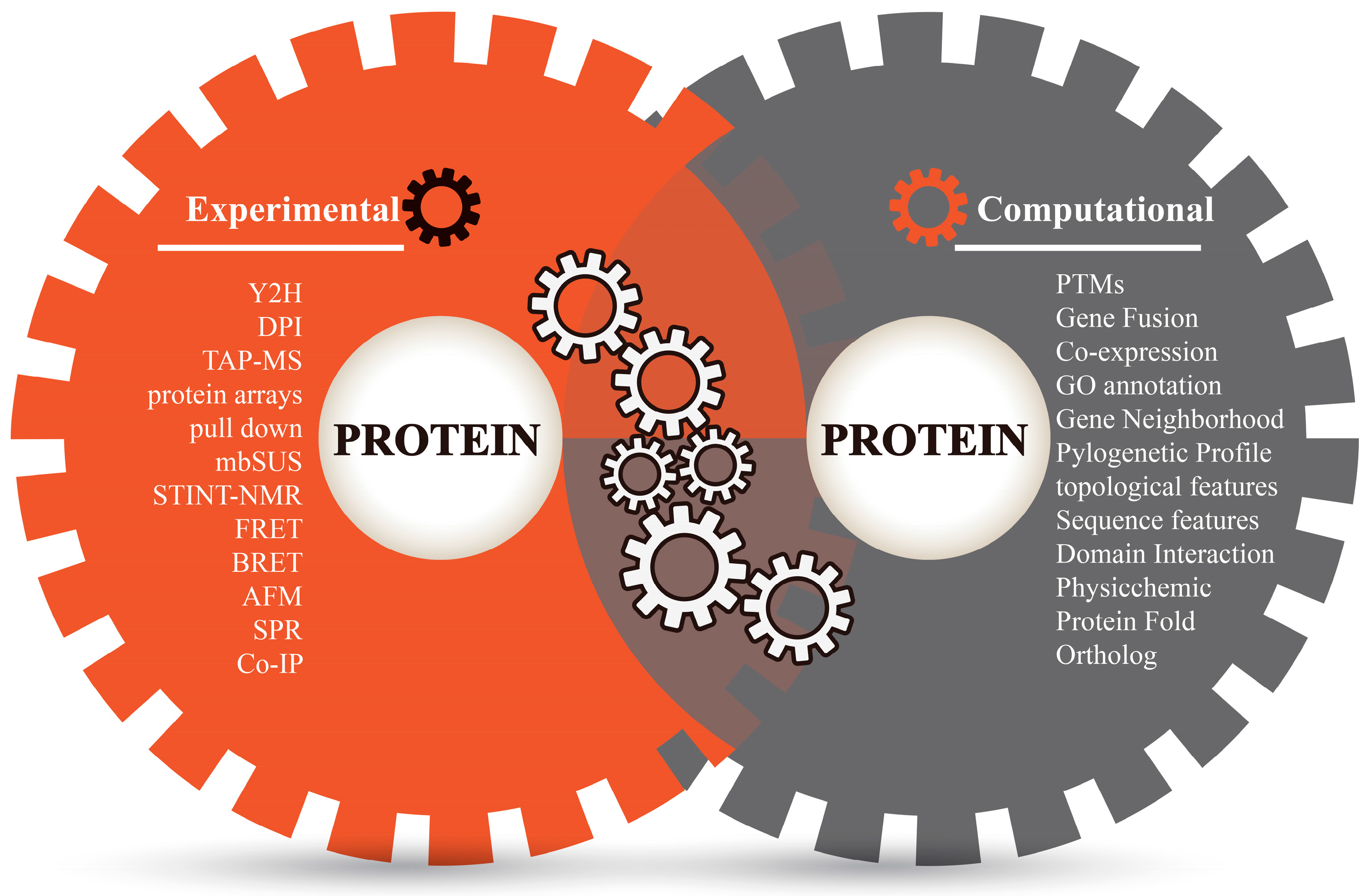
Recent advances in predicting and modeling protein–protein interactions: Trends in Biochemical Sciences

Scheme for all-to-all protein-protein interaction predictions. Using a... | Download Scientific Diagram

BindML: Software for Predicting Protein-Protein Interaction Sites using Phylogenetic Substitution Models
![PDF] ELASPIC web-server: proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity | Semantic Scholar PDF] ELASPIC web-server: proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/94352efe483329f00e3e77e0922562f7bc98b78b/2-Figure1-1.png)
PDF] ELASPIC web-server: proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity | Semantic Scholar





![PDF] The FunFOLD 2 server for the prediction of protein ligand interactions | Semantic Scholar PDF] The FunFOLD 2 server for the prediction of protein ligand interactions | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/c50225f129c18e56d2cbe4352383ce46df326d70/5-Figure1-1.png)